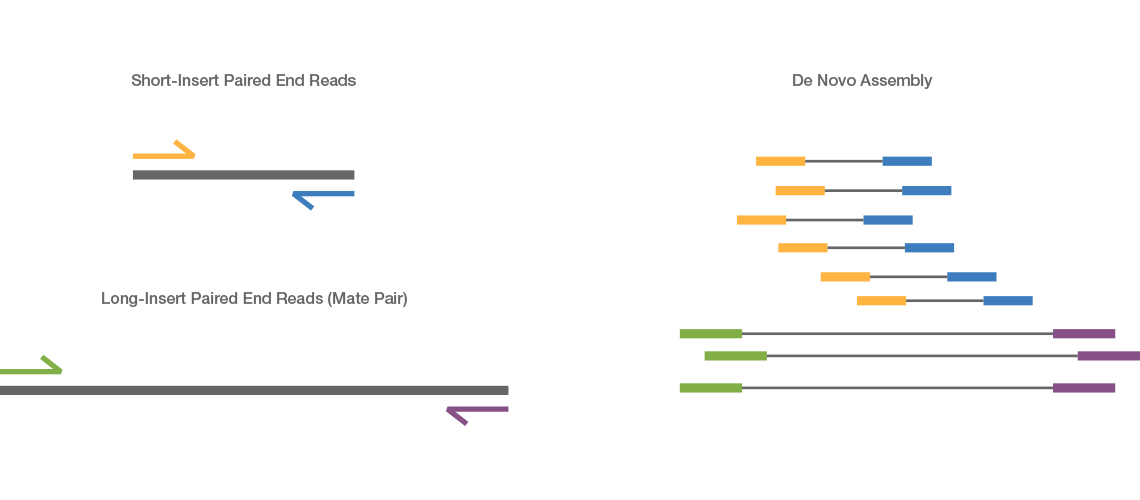
paired end sequencing reads
Paired-end DNA sequencing reads provide high-quality alignment across DNA regions containing repetitive sequences and produce long contigs for de novo. When you align them to the genome one read should align to the forward strand and the other should align to the reverse strand at a higher base pair position than the first one.

What Are Paired End Reads The Sequencing Center
Since paired-end reads are more likely.

. In theory paired end reads should not overlap. For sequencing projects that require higher accuracy such as studies of alternate splicing 40 million to 60 million paired-end reads will provide better results. The standard Illumina paired-end protocol produces reads oriented pointing toward each other just like good old fashioned Sanger paired reads but the insert size is much.
Paired-end vs single-end sequencing reads. For more detailed analyses to. Visit Maverix Biomics to learn more about RNA-seq.
TW-201243117-A chemical patent summary. Paired-end sequencing facilitates detection of genomic rearrangements and repetitive sequence elements as well as gene fusions and novel transcripts. Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data.
The Illumina paired-end sequencing technology can generate reads from both ends of target DNA fragments which can subsequently be merged to increase the. Mate pair sequencing is used for various applications applications including. Paired-end sequencing facilitates detection of genomic.
In general paired-end reads tend to be in a FR orientation have relatively small inserts 300 - 500 bp and are particularly useful for the sequencing of fragments that. By merging paired-end reads the overlapping region between them can also be deployed for correcting. Paired-end DNA sequencing reads provide high-quality alignment across DNA regions containing repetitive sequences and produce long contigs for de novo sequencing by filling gaps in the.
Should you merge paired end reads. Combining data from mate pair sequencing with those from short-insert paired-end reads provides increased. With paired-end sequencing after a DNA fragment is read from one end the process starts again in the other direction.
The Illumina paired-end sequencing technology can generate reads from both ends of target DNA fragments which can subsequently be merged to increase the. RNA-seq analysis configuration on the Maverix Analytic Platform. Biocc paired end or mate pair refers to how the library is made and then how it is sequenced.
This can be done using the Set Paired Reads. Both are methodologies that in addition to the sequence information give you. In addition to producing twice the number of sequencing reads this.
Read files from paired-end sequencing need to be paired in Geneious before the pairing information can be used in assembly.
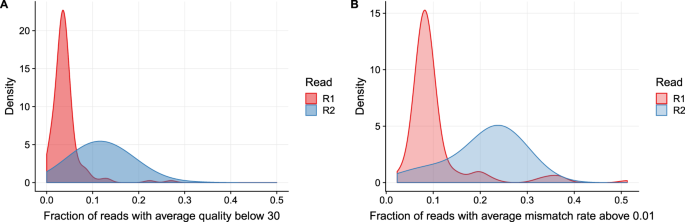
Long Fragments Achieve Lower Base Quality In Illumina Paired End Sequencing Scientific Reports
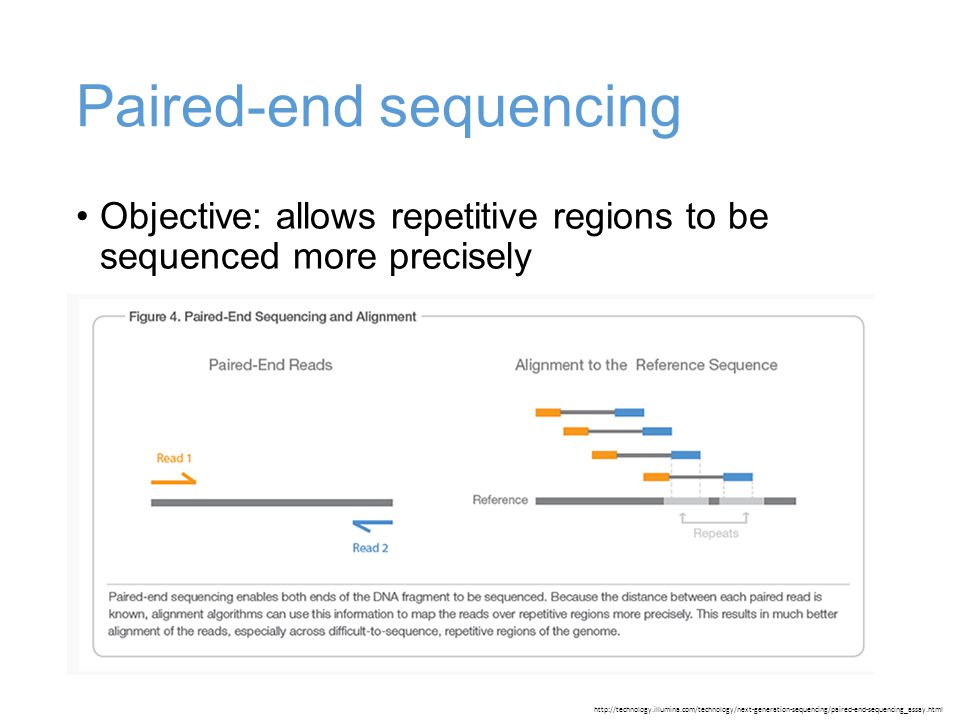
Mcb Lecture 9 Sept 23 14 Illumina Library Preparation De Novo Genome Assembly Ppt Download
How Sequencing Works Ngs Analysis

Paired End Sequencing Rna Seq Blog
![]()
Overview Of Amplicon Preprocessing
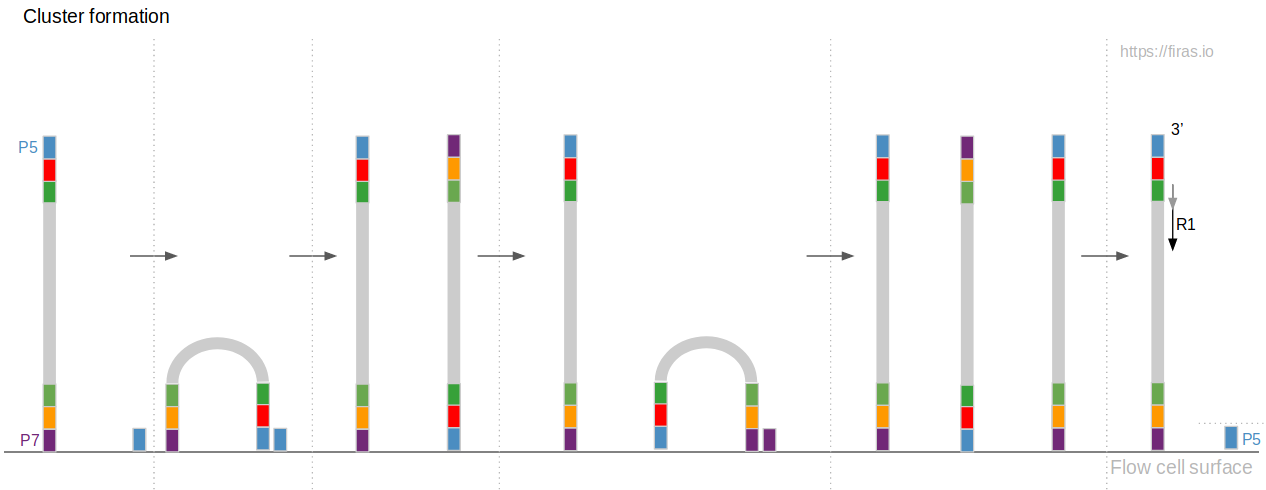
An Overview Of Illumina Multiplexing Firas Sadiyah
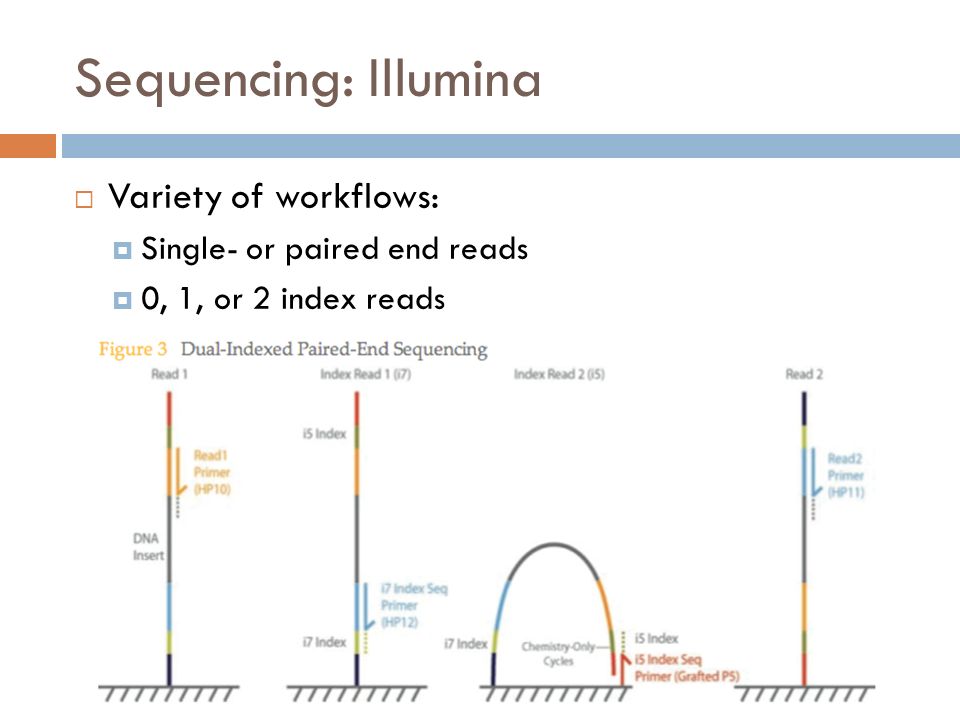
Experimental Design And Sample Preparation Vmi Bootcamp Ii 1 0 Documentation

Paired End Sequencing E Learning Vib

Beyond The Linear Genome Paired End Sequencing As A Biophysical Tool Trends In Cell Biology

Long Fragments Achieve Lower Base Quality In Illumina Paired End Sequencing Biorxiv
Personal Genome Sequencing Current Approaches And Challenges
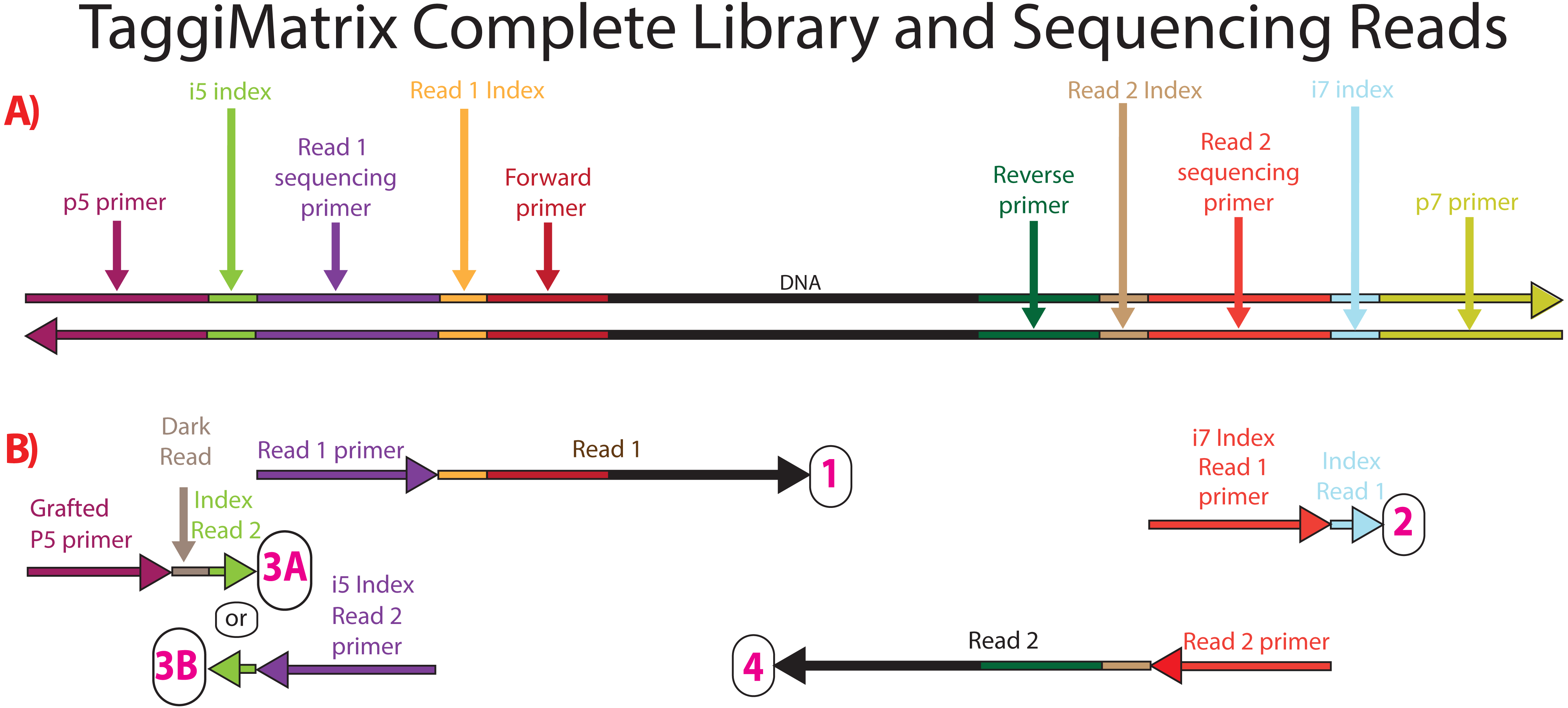
Adapterama Ii Universal Amplicon Sequencing On Illumina Platforms Taggimatrix Peerj

Schematic Representation Of Paired End Sequencing For Very Short Download Scientific Diagram

Rna Seq Data Analysis 1 Raw Single End And Paired End Reads Obtained Download Scientific Diagram

Orientation Of Pe Reads A Review Of Fr Ff And Rf Meanings
How Sequencing Works Ngs Analysis
Local De Novo Assembly Of Rad Paired End Contigs Using Short Sequencing Reads Plos One